 "Technical
Skill is mastery of complexity while creativity is mastery of simplicity"
"Technical
Skill is mastery of complexity while creativity is mastery of simplicity" "Technical
Skill is mastery of complexity while creativity is mastery of simplicity"
"Technical
Skill is mastery of complexity while creativity is mastery of simplicity"
I. Clinical Chemistry
History of Cardiac Troponin I in Clinical Chemistry
The first commercially available Troponin I Immunoassay was developed at Dade International (aka: Baxter Diagnostics, Inc/Dade-Behring, Inc) , Miami, Florida back in the summer of 1995. I was fortunate enough to be part of this exciting project. I worked on the stabilization and optimization of cardiac Troponin I and the actual development of this test. It was an exciting 4.5 years that took to complete the project and get expedited review from the FDA. The assay gave the company an edge for about 3 years before competitors realized that they had fallen behind in the cardiac markers arena. Baxter always enjoyed a great position as they also had one of the first CKMB mass assays in the market. Troponin was the next generation of the cardiac assays. Anyways enjoy the reading. Upon completion of the project the USPTO awarded the scientist that worked on this assay a patent for some of the work involved. (See documents)
Troponin I Historical Documents (Check out the FDA Documents fromthe original release of the first assay in the US and Europe)
Troponin I is a protein found in both skeletal
and cardiac (heart) muscle. It is part of the ternary complex of tropomyosin.
These are the proteins that act as a chemical switch involved in the calcium
dependent contraction of muscles. The other two proteins associated with the
tropomyosin complex are: cardiac troponin C (calcium binding) and T
(tropomyosin). In the late 1980's a British scientist published a scientific
paper on the potential utility of cardiac troponin I as a
marker for myocardial infarction (MI). At that time I was a scientist in
industry
and was in a research group developing cardiac marker immunoassays. We were in charge of developing the chemistry of these kits to go
along with the expensive instrumentation designed to analyze serum samples for this protein. As a researcher
in industry we teamed up with a Washington University School of Medicine
and
started a project in 1991 at Dade International (a.k.a.Baxter, Dade-Behring), to develop a sandwich
immunoassay to detect and quantitate the presence of this protein in serum.
Interestingly enough a group of three folks (inclusive of yours truly) worked
feverishly for about 4 years to learn how to stabilize and prevent this protein
from degradation.
A
patent was granted for some of the work involved in stabilizing this protein in
solution. Cardiac troponin I is a very basic and not very soluble protein.
We
needed to stabilize troponin I in order to develop standard solutions that can be stored refrigerated or
lyophilized and used without significant changes in values assigned. The FDA
requires that these medical devices have certain stability or shelf life before
they are approved for sale. Needless to say once the kit was developed and
serious challenges were overcome the clinical trials were performed to determine
the clinical sensitivity and specificity of this test in the detection of
myocardial infarction. It was found after analyses of the clinical data that
cardiac troponin I is a very specific (selective) marker for myocardial
infarction. The trick is to have antibodies developed for an epitope (amino acid
sequence) that is unique to the cardiac and not the skeletal Troponin I. Thus, the immunoassay will
detect Troponin I in serum originating from cardiac muscle. The sensitivity was excellent specially after 12 hours (100%)
post MI event. Cardiac troponin I becomes elevated in folks with MI and can
be detected after 4 hours post MI incident. Normal folks who are not
undergoing heart attack will have essentially undetectable levels of this
protein in their serum. Of course now the assay's sensitivity is much better and
the low level numbers which are not MI related appears to have risk
stratification value. TnI remains elevated for about a week following the heart
attack which is useful for folks that wait to be seen by their doctors. In folks with unstable angina
slightly elevated levels of cTnI have been reported. As you all may know doctors
do not use serum markers exclusively, but in conjunction with ECG
(electrocardiogram), clinical history, and other serum markers too. The
serial testing of cTnI is essential to make sure the values take off and clear
the 99th percentile of "normals". Cardiac Troponin I has helped send more chest pain patients home by ruling out heart attack based on a couple of serial bleeds. Therefore, these patients
do not need to be admitted or stay beyond 24 hrs in an ED taking up
expensive beds and resources. The cost savings is in the billions. The more
sensitive a test the less likely it is that a physician send a patient home with
an MI.
II. Cardiac Troponin I as a Time of Death Marker
After departing with Dade Bering which reIocated to Delaware, I returned to FIU to start my PhD in Chemistry. I had certain interests in researching the utility of cardiac Troponin I as a time of death marker. A time of death marker is a protein, chemical, or parameter that can be used by a forensic pathologist to estimate the postmortem interval or time since death. In television folks use core body temperature as a time of death estimator. Although it is true that forensic pathologist use its limitations are numerous. I embarked on developing of a technique to extract cTnI from human cardiac tissue and characterized the rate of degradation of the protein postmortem.
Purification of TnC and TnI
In the gel above we find cTnI purified by
affinity Sepharose-TnC because of the physiological interaction between cardiac
TnC and cTnI in the presence of calcium the two molecules associate with a high
affinity constant and in the absence (chelator) they dissociate (a chemical switch).
Troponin C was purified by ion exchange chromatography and
the Troponin I by affinity chromatography using covalently immobilized Troponin C.
Determination of molecular mass (weight) can be performed by plotting the
migration distance versus the log of the molecular weight of the standard
protein. As shown by the graph to the right.
In another words, can this protein be used to predict the time since death of a cadaver? The results were so interesting that a patent application was applied for a few years back.. Our first publication was accepted by the Journal of Forensic Science International.
Purification of Monoclonal Antibodies
For this research I purified some
monoclonal antibodies (IgG) from ascites
fluid by a protein G column. Protein G column has affinity for the Fc
region of the Immunoglobulin G (IgG) antibody. It binds with a high affinity
constant and then the unbound proteins are washed and eluted from the column at a low pH
buffer. A coomassie gel is run to verify the presence of these proteins under
denaturing/reducing conditions. IgG are antibodies developed against specific  antigens
and thru hybridoma technology to obtain a single cell line called a
monoclonal (one clone) that is very consistent and reproducible. Today we no
longer need to use mice to raise and grow the antibody. Bioreactors
can be used to generate large quantities of these clones used in applications
such as therapeutics, diagnostics, etc. IgG (~150 ka is composed of two heavy and two
light chains that separate under SDS-PAGE reducing conditions. IgG are the same
proteins that confer our humoral immunity. These proteins are
rugged and not very thermally labile given their physiological function. As can
be seen is the gel to the right: the ascites contains other proteins found in
ascites fluid which are eliminated in the affinity purification.
antigens
and thru hybridoma technology to obtain a single cell line called a
monoclonal (one clone) that is very consistent and reproducible. Today we no
longer need to use mice to raise and grow the antibody. Bioreactors
can be used to generate large quantities of these clones used in applications
such as therapeutics, diagnostics, etc. IgG (~150 ka is composed of two heavy and two
light chains that separate under SDS-PAGE reducing conditions. IgG are the same
proteins that confer our humoral immunity. These proteins are
rugged and not very thermally labile given their physiological function. As can
be seen is the gel to the right: the ascites contains other proteins found in
ascites fluid which are eliminated in the affinity purification.
The chromatogram (left below) shows the loading, washing and elution of the
antibody from a protein G column. The antibody elute at pH 2.7 must be quickly
neutralized with tris buffer pH 9 to avoid denaturation of the anitbody and
precipitation. The antibody must be buffered exchanged by dialysis or amicon to
a PBS solution.
Western Blot
A Western blot is a technique of transferring
proteins from a gel where they cannot be truly probed with antibodies to a paper
that binds proteins. This process can be allowed to 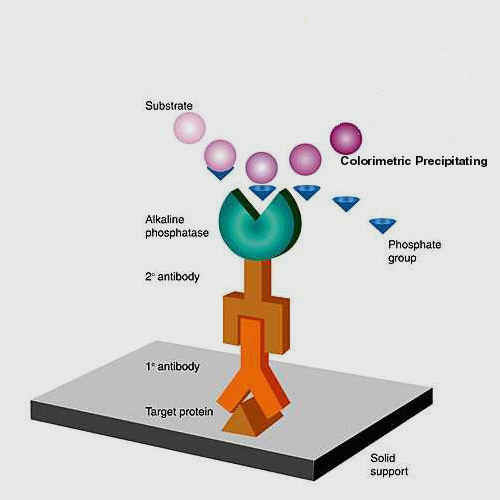 happen
by simple diffusion from the gel to the paper (high to low concentration). The
process is much more efficient if we electroblot the protein using buffers that
help the proteins elute from the gel and adhere to the paper as a current
is passed across a cathode and anode. Once transferred, the paper the paper is
blocked with milk protein (casein) and then the proteins can be probed with
antibodies directed at different epitopes of the protein. Once the antibody
binds a conjugate consisting of a class capture goat anti mouse (GAM-ALP or GAM-
HRP) antibody linked to an enzyme can be used to bind the mouse antibody bound
on the paper. The membrane is washed at every step and the a substrate that
becomes insoluble in the presence of alkaline phosphatase (ALP) is used to
visualized the bands with the protein of interest.
happen
by simple diffusion from the gel to the paper (high to low concentration). The
process is much more efficient if we electroblot the protein using buffers that
help the proteins elute from the gel and adhere to the paper as a current
is passed across a cathode and anode. Once transferred, the paper the paper is
blocked with milk protein (casein) and then the proteins can be probed with
antibodies directed at different epitopes of the protein. Once the antibody
binds a conjugate consisting of a class capture goat anti mouse (GAM-ALP or GAM-
HRP) antibody linked to an enzyme can be used to bind the mouse antibody bound
on the paper. The membrane is washed at every step and the a substrate that
becomes insoluble in the presence of alkaline phosphatase (ALP) is used to
visualized the bands with the protein of interest.  Below
is a western blot of bovine cardiac troponin I. These blots can detect down to
low picogram levels of the protein.
Below
is a western blot of bovine cardiac troponin I. These blots can detect down to
low picogram levels of the protein.
SIZE EXCLUSION CHROMATOGRPAHY
As part of my research I performed some size
exclusion chromatography (SEC) (a.k.a gel filtration or gel permeation
chromatography). In this type of chromatography the proteins are separated by a
resin that has a  given
size exclusion limit. You choose the resin appropriate for the molecular mass
separation desired, In the case of the proteins, I used a Pharmacia Superose-6
column for the FPLC (Fast Protein Liquid Chromatograph) separation of some
proteins and to see if I could detect aprotinin in a proprietary formulation of
protease inhibitors. The use of SEC in separation of macromolecules is useful
although many times it is not feasible to obtain baseline resolution in your
separation because you can collect fractions and the area of overlap can be isolated
and separated once again. The alternative is to use a narrower longer
column and of course that is not so practical in the real world. Typically gel
permeation is the term used by those who separate polymers by SEC with
non-aqueous solvents and gel filtration is reserved for the separation involving
aqueous mobile phase.
given
size exclusion limit. You choose the resin appropriate for the molecular mass
separation desired, In the case of the proteins, I used a Pharmacia Superose-6
column for the FPLC (Fast Protein Liquid Chromatograph) separation of some
proteins and to see if I could detect aprotinin in a proprietary formulation of
protease inhibitors. The use of SEC in separation of macromolecules is useful
although many times it is not feasible to obtain baseline resolution in your
separation because you can collect fractions and the area of overlap can be isolated
and separated once again. The alternative is to use a narrower longer
column and of course that is not so practical in the real world. Typically gel
permeation is the term used by those who separate polymers by SEC with
non-aqueous solvents and gel filtration is reserved for the separation involving
aqueous mobile phase.
Design of Experiments (DOE)
As part of my research, I also performed several experiments that involved optimization of an ELISA (Enzyme Linked Immunosorbent Assay). This ELISA was to geared to measure quantitatively cardiac Troponin I. The optimal fashion to perform experiments is to change several factors (variables) that affect an output (response) at the same time. Design of experiments is the use of multivariable regression to fit a mathematical model to experimental data that dictate different levels of a factor in combination with other factors. In this manner we are changing several variables in one experiment and looking for interactions with other variables. The mathematical models are diverse and a typical starting point is a linear screening experiments that measures the main or primary effects of the factors, The data below was generated with a response surface methodology (RSM) following a quadratic model that can fit nonlinear relationships and obviously find maximas and minimas. The experiments are performed in a random fashion and a set of conditions are repeated throughout to obtain information on the precision of the technique. The matrix or worksheet of conditions to run is sometimes too costly in time and money so other models such as fractional factorials can be used to obtain information on the effects. The less conditions tested the chances for confounding effects in the data generated, Typically a model that can discriminate first and second order interactions is sufficient for most scientists. Third order interactions are rare and are not as important as first and second order. The traditional approach in science has been to change one variable at a time and measure its effects. The design of experiments approach is to vary several important variables and see if the response obtained is actually non-linear. If you apply a linear model to a second order interaction you may potentially miss a maxima or minima because of the fitting routine. You also perform analysis of variance (ANOVA) to compare the different conditions. A high residuals may be indicative of a poor fitting routine (lack of fit) and may indicate the use of a second order model. Although there is great resistance to DOE in some sectors of the scientific community many times scientists who realize the importance and value of it will never again resort to single variable experiments, unless, the nature and cost of experimentation is prohibitive. The FDA is currently is requiring research groups in companies to follow design control techniques whereby scientists have to develop products and formulations by using experimental design to show the scientific reasoning behind a given formulation. The statistical basis for these experiments lends lots of credibility to the outcomes and conclusions. Not to mention that challenges to patents or formulations may be justified based on empirical data analyzed and predictive of a given formulation.



If you have any questions please feel free to contact me.